DNA Nanotechnology
DNA is the carrier of hereditary information and therefore a central component of all life on earth. However, due to its unique chemical and structural properties, DNA can also be used as a programmable material for the controlled synthesis of molecularly defined artificial nanostructures. The DNA origami technique enables the fast, high-yield synthesis of 2D and 3D nanostructures by exploiting the strong specificity of Watson-Crick base pairing. It is based on the folding of a long single-stranded DNA scaffold into a desired shape with the help of hundreds of short synthetic oligonucleotides. This technique provides a straightforward means to assemble user-defined nanostructures with arbitrary, yet well-defined shapes. Furthermore, the unique sequences of the oligonucleotides employed in DNA origami folding enable the resulting DNA origami nanostructures to serve as spatially addressable molecular breadboards for the controlled arrangement of biomolecules and nanoparticles with nanometer precision. We are exploring potential real-world applications of DNA origami technology in various application areas.
DNA nanostructure stability
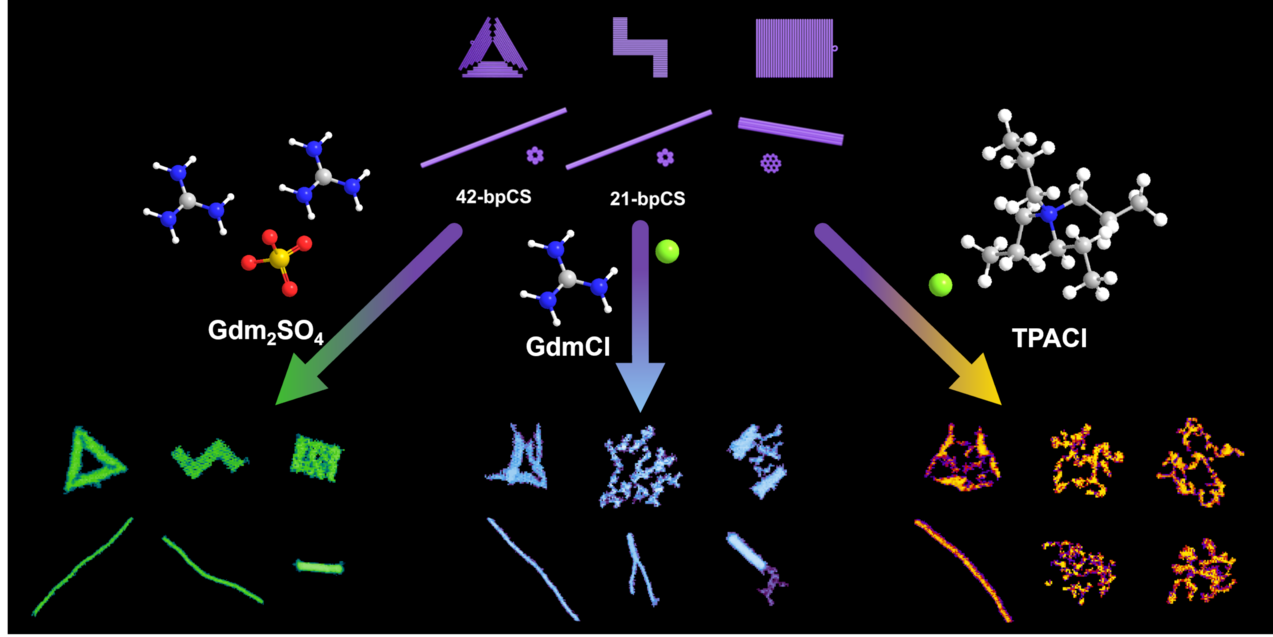
Many applications of DNA origami nanostructures in medicine, technology, and research rely on an intact and well-defined DNA origami shape, for example to present functional molecules in well-defined geometric arrangements. In such instances, thermal fluctuations and limited stability under relevant conditions often become critical factors. This is further complicated by the fact that DNA origami nanostructures present some unique structural properties including numerous non-natural DNA topologies that may deviate substantially from genomic DNA. We, therefore, investigate the stability of DNA origami nanostructures under various relevant conditions with special emphasis on the impact of molecular interactions and design parameters.
Selected publications:
- Superstructure-dependent stability of DNA origami nanostructures in the presence of chaotropic denaturants, M. Hanke, D. Dornbusch, E. Tomm, G. Grundmeier, K. Fahmy, and A. Keller, Nanoscale 15 (2023) 16590 (Cover Picture)
- Effect of Ionic Strength on the Thermal Stability of DNA Origami Nanostructures, M. Hanke, E. Tomm, G. Grundmeier, and A. Keller, ChemBioChem 24 (2023) e2023003
DNA-based digital data storage


Humanity is producing and storing data at an unprecedented rate. Unfortunately, all current storage media have fundamental limitations when it comes to long-term archiving, particularly in the form of scaling problems and constant media obsolescence, which means that the stored data must be migrated on a regular basis. Current storage technologies are thus too costly and also have significant negative environmental impacts. We are part of an interdisciplinary international consortium funded within the framework of the EIC Pathfinder project "Next Generation Molecular Data Storage (NEO)", which explores the possibility to write digital data as molecular bits in DNA origami nanostructures, read them out by atomic force microscopy, and reconstruct the original data with the help of computer vision and machine learning approaches. Our sub-project in particular is focussed on the development of data-carrying DNA origami nanostructures with maximum storage density and long-term stability in dry and liquid environments.
Selected publications:
- Cost-efficient folding of functionalized DNA origami nanostructures via staple recycling, E. Tomm, G. Grundmeier, and A. Keller, Nanoscale 17 (2025) 17265
- Toward high-density streptavidin arrays on DNA origami nanostructures, L. Rabbe, E. Tomm, G. Grundmeier, and A. Keller, RSC Adv. 15 (2025) 24536
DNA-controlled enzyme reactions

The control of enzyme reactions is of extraordinary importance for diverse areas in fundamental and applied research such as (nano)biotechnology, synthetic biology, and biomedicine. In the context of DNA origami, enzyme reactions are of particular relevance for the structural integrity of the DNA nanostructures in adverse environments. In physiological media, for instance, nucleases digest DNA origami nanostructures and thereby limit their in-vivo lifetime. Ligases, on the other hand, can be used to seal discontinuities in the DNA backbone, so-called nicks, and thereby increase the thermal stability of DNA origami nanostructures. Both effects are critically influenced by DNA origami superstructure, both on a local and a global level. We thus investigate the effects of various structural and design parameters on ligase and nuclease reactions in different DNA origami nanostructures with the aim to develop general design rules for controlling enzyme reactions in these nanostructures.
Selected publications:
- Environment-Dependent Stability and Mechanical Properties of DNA Origami Six-Helix Bundles with Different Crossover Spacings, Y. Xin, P. Piskunen, A. Suma, C. Li, H. Ijäs, S. Ojasalo, I. Seitz, M.A. Kostiainen, G. Grundmeier, V. Linko, and A. Keller, Small 18 (2022) 2107393
- Real‐Time Observation of Superstructure‐Dependent DNA Origami Digestion by DNase I Using High‐Speed Atomic Force Microscopy, S. Ramakrishnan, B. Shen, M. A. Kostiainen, G. Grundmeier, A. Keller, and V. Linko, ChemBioChem 20 (2019) 2818 (Cover Picture)
DNA-based nanofabrication

Many potential applications of DNA origami nanostructures in (bio)chemical sensing, lab-on-chip diagnostics, and tissue engineering, rely on their deposition on solid substrates. Furthermore, DNA origami nanostructures immobilized on solid surfaces can also be used as molecular lithography masks to transfer their shapes into various organic and inorganic materials. Therefore, the controlled deposition of single DNA origami nanostructures, as well as their hierarchical self-assembly into ordered 2D lattices represent important steps toward such real-world applications. We thus investigate DNA origami adsorption and lattice formation on solid substrates to elucidate the underlying molecular mechanisms and optimize the size and quality of the lattices. We furthermore utilize adsorbed DNA origami nanostructures for surface functionalization using molecular lithography techniques.
Selected publications:
- On the role of cation-DNA interactions in surface-assisted DNA lattice assembly, X. Xu, B.K. Pothineni, G. Grundmeier, S. Tsushima, and A. Keller, Nanoscale (2025)
- Cation-dependent assembly of hexagonal DNA origami lattices on SiO2 surfaces, B.K. Pothineni, G. Grundmeier, and A. Keller, Nanoscale 15 (2023) 12894 (Cover Picture)

